Software
ctyper
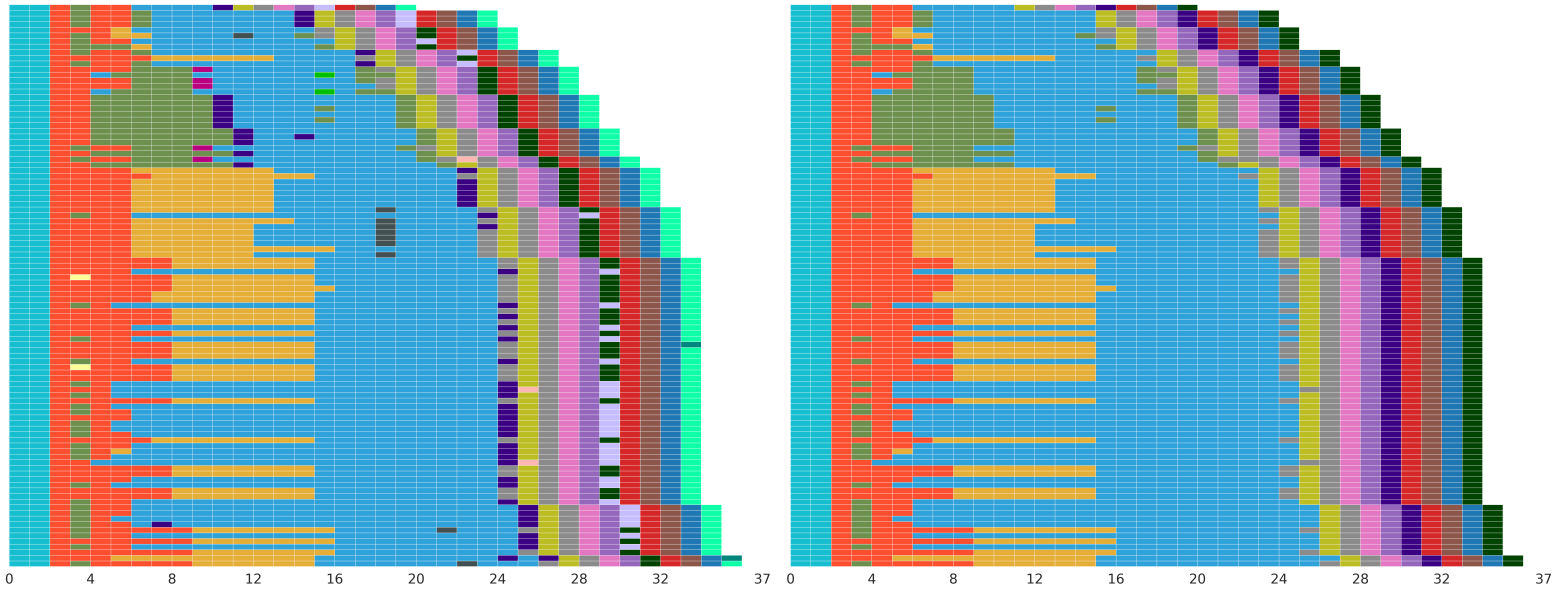
Ctyper is a genotyping approach for pangenomes to detect alleles that are shared between a biobank (short-read sequencing) sample and a long-read pangenome for challenging and copy-number variable regions of the genome.
bioconda: ctyper
git: https://github.com/chaissonlab/ctyper
publication: https://www.nature.com/articles/s41588-025-02346-4
author: Walfred Ma
vamos
STR and VNTR annotation by motif composition for PacBio and Oxford Nanopore reads, and assemblies. The vamos suite contains two components, one to characterize the repeat motifs in a population using an efficient motif set, a subset of motifs that preserves motif sequence diversity while removing rare motifs, and annotation software that uses wraparound dynamic programming to annotate samples using tandem repeat and motif databases. bioconda: vamos
git: https://github.com/chaissonlab/vamos publication: https://link.springer.com/article/10.1186/s13059-023-03010-y
authors: Jingwen Ren and Bida Gu
Owl
Detects microsatellite instability in PacBio HiFi reads.
git: https://github.com/pacificbiosciences/owl/
author: Zev Kronenberg
LRA
Software for mapping long reads (PacBio/Oxford Nanopore) or their assemblies to genomes. LRA implements an exact convex gap penalty for biologically meaningful gaps to discover structural variation.
git: https://github.com/chaissonlab/lra
bioconda: lra
publication: https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1009078
author: Jingwen Ren
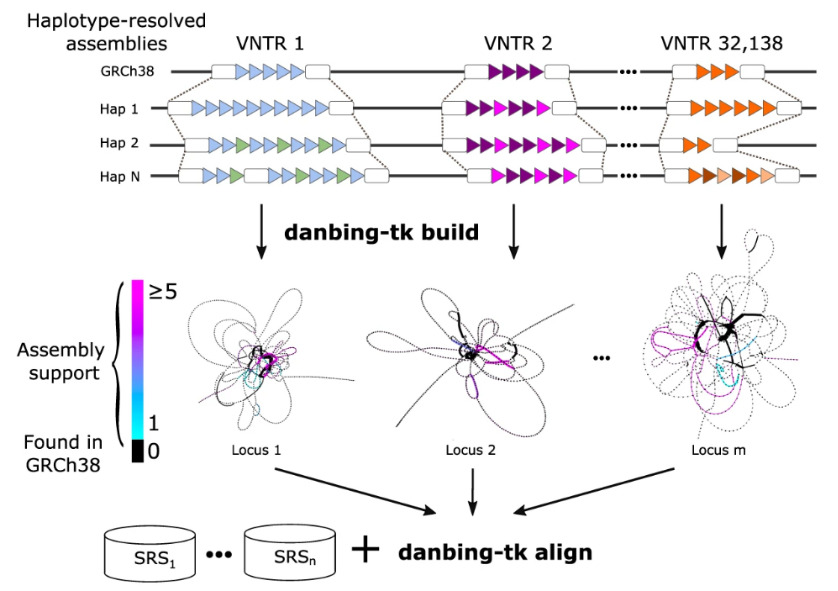
danbing-tk
danbing-tk is a suite of tools to construct locus-specific de Bruijn graphs on tandem repeat loci from pangenomes (repeat-pangenome graphs), and to map short reads to the graphs. Length and motif variation can be inferred from alignment depth.
git: https://github.com/chaissonlab/danbing-tk
publications: https://genome.cshlp.org/content/33/4/511.short, https://www.nature.com/articles/s41467-021-24378-0
author: Tony Tsung-Yu Lu